On March 28, 2018 has been released a new version of GCDkit 5.0. It was developed in R 3.4.3 and code-named El Chupacabra. The most important changes are summarised below.
Towards a platform-independent version
- It is well-known that the GCDkit has been originally designed as a Windows-only package. From the current version it is possible to install and run it also on operation systems other than Windows, most notably Linux or Mac OSX.
- So far, this concerns command-line/batch mode only.
- The platform-independent graphical user interface (menus, dialogue boxes...) written in Tcl/Tk language is ready and just being tested. If you are feeling unadventurous, and work on Linux and/or Mac, please do get in touch and you can become one of the beta testers.
New Windows installer
- As we hope, we have fixed the long-standing problems with the installation of GCDkit on Windows 10, and people without administrative rights.
- The new GUI installer, based on the Tcl/Tk language, is started by simply dragging the file @INSTALL.r from the temporary installation directory onto the R Console window.
- his file can be also located manually and loaded using the File|Source R code...' menu. See the download page for more details.
- Please note that this installer relies on a live Internet connection to download the missing packages required by the GCDkit.
User-defined templates
- User-defined templates for stand-alone Figaro plots or their plates. They can be uploaded into the directory Diagrams/User and then employed for standard plotting (i.e. not classification) purposes. This mechanism allows easy and fully automatic expansion of the plotting capabilities of the GCDkit system. For further info, see the file @README.txt in the directory in question.
- There are five examples of user-defined templates provided: three binary plots for discrimination of lavas from destructive plate boundaries (Pearce 1982), ternary plot for classification of A-type granites (Grebennikov 2014) and binary plot of Paulick et al. (2006) to portray the mantle peridotite fertility.
- Functions allowing overplotting new datapoints onto standard single binary plots, ternary plots,
spiderplots or multiple plots, including the diagram templates (overplotDataset, underplotDataset) linked to the menu system. - In addition, there are functions figOverplot and figOverplotDiagram but these are meant mostly for internal use.
- For plotting serves a reference dataset, either real-world data (say compiled from literature) or a numeric matrix spanning, for instance, from petrogenetic modelling.
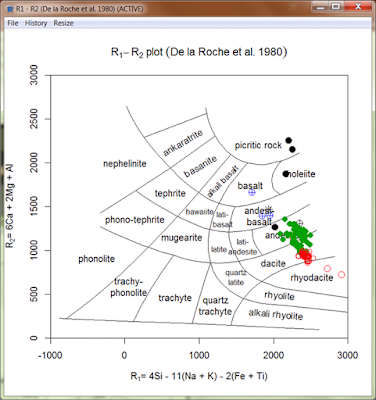
- See the example of overplotting the atacazo over sazava dataset on De la Roche et al. (1980) plot.
- Very attractive feature should be the new possibility of obtaining dataset by online search of the EarthChem.org database. The search can be performed directly from the menu GCDkit|Online search EarthChem.org. It invokes a graphical front-end, written By Oscar Laurent (ETH, Zurich) that allows to enter the desired search criteria. On Windows, it also includes a primitive, and still rather slow, possibility of selecting a polygon of interest from a map of the World.
Further new features/important changes
- New plugin 'disclosure' for log-transformation of compositional data.
- New geotectonic/general purpose diagrams
- La/Yb vs. Nb/La and La/Yb vs. Th/Nb of Hollocher et al. (2012)
- La/10-Y/15-Nb/8 of Cabanis and Lecolle (1989)
- Zr/Y-Th/Yb of Ross and Bedard (2009)
- YbN vs. LaN/YbN of Martin (1986) to distinguish adakites and TTGs
- F-M-W diagram (Ohta and Arai 2007) for chemical weathering of igneous rocks
- calcAnomaly, a function for calculating a magnitude of any anomaly on any spiderplot, based on concentrations of selected neighbouring (not necessarily adjacent) elements.
- Spanish translation of the classification plots (courtesy of Tomas Grijalva, Geology Department of UNAM, Mexico).
- New zircon saturation model of Boehnke et al. (2013).
- New training dataset atacazo.data giving the whole-rock major- and trace-element contents, together with Sr and Nd isotopic compositions of lavas from two volcanic complexes in Ecuador: the Atacazo and the Ninahuilca (Hidalgo 2006; Hidalgo et al. 2008). This dataset, kindly provided by Silvana Hidalgo, is used in a worked example (chapter 25) of Janousek et al.’s book (2016).
- Another new training dataset blatna.data from the Central Bohemian Plutonic Complex, Czech Republic (Janousek et al. 2000, 2010). It could be, together with sazava.data already present, used to test dataset switching etc.
- New function figFixLim() extends the scales of both axes of a binary plot automatically if necessary to accommodate all the data points.
- new training dataset blatna.data from the Central Bohemian Plutonic Complex, Czech Republic (Janousek et al. 2000, 2010). It could be, together with sazava.data already present, used to test dataset switching etc.
- New function figFixLim()extends the scales of both axes of a binary plot automatically if necessary to accommodate all the data points.
- New normalization scheme of Anders and Grevesse (1989) for chondrite-normalized REE spiderplots.
- Better behaviour of many functions in batch mode (in RTerm, Jupyter, on Linux/Mac etc.)......
We trust that you will find this new release useful and reasonably stable.
Good luck, Vojtech





Hello ... can anyone help me?
ReplyDeleteI have been using CGDkit for some time and I can not install the new version in Windows 10, because when I unzip the folder, it does not have the Run RGUI file, 32 bit version (shortcut labeled R i386).
Hello, yes, the installation philosophy has changed completely - and you should read Instructions at our download page, http://www.gcdkit.org/download. Have you installed R first? GCDkit is just a library for R.
DeleteHello, thank you for replying. Yes, I have read and followed the instructions. The R was installed, however from "Run RGUI, 32 bit version (shortcut labeled R i386)" was not possible.
DeleteHello,
Deleteto be concerete, the shortcut to 32-bit R on my desktop is named R i386 3.4.3. Can you find it and start it?
Hi!
ReplyDeleteThe new version of the GCDkit is really nice but I can't select the elements when I want to do a spider plot. There is another way to do it in this new version? In the previous GCDkit versions it was possible to do it.
Cheers!
Hello,
ReplyDeleteI am not sure what are you after - if it is introducing your own normalizing scheme for spiderplot, or modifying an existing one, please refer to:
http://blog.gcdkit.org/2013/06/how-to-introduce-your-own-normalization.html
Or please provide more details.
Hi,
DeleteSorry for the late reply. That link really helped.
Thank you for the help.
This comment has been removed by the author.
ReplyDeleteHi! Thanks for all the tips in the blog. They really help. I was used to GCDkit in PC, but now I managed to install XQuartz, R 3.5.1, the packages ("R2HTML","RODBC","XML","sp") and load GCDkit on HighSierra 10.13.6. When I load the library with library(GCDkit) there is an error message:
ReplyDeleteError reading the GCDkit config file!
Sticking to defaults....
=====================================
Patching.... please wait
Initializing the EarthChem interface....
Testing - on Non-Windows system - skipping Eartchem interface
#but after it, there is the normal GCDkit message followed by:
"Ready 2 go - Enjoy!"
I set the working directory, but when I try to loadData, if it is an .xlsx file, the following message appears:
Excel 2007 file...WARNING: MS Excel import is not available on 64bit systems! Quitting...
File reading error
#.xls files also do not load
#And if I try a .csv file, the message is:
CSV file...loading...
Error in file(file, "rt") : não é possível abrir a conexão
File reading error
Error in sqlQuery(channel, paste("SELECT * FROM [", which.table, "]", :
objeto 'channel' não encontrado
Is there any component I didn't install? If it is the case to run R in 32 bits, how can I change it in ios?
Thanks
Ola Isabela,
Deleteobrigadinho for your bug report. It seems that there are some problems with the RODBC library used to read Excel and CSV files. I am not working on Mac but I suspect that the ODBC drivers are available only on Windows, and then only on 32 bit systems. To be on the safe side, you can stick to text files (just copy and paste from you Excel). Meanwhile, let us to examine the problem further.
Cheers,
Vojtech
Olá Vojtech, thank you for your reply!
DeleteI also tried GCDkit 4.1 in R 3.2.1 and the error messages are:
GCDkit-> loadData("teste1.xlsx")
Excel 2007 file...Error in try(switch(suffix, ROC = { :
não foi possível encontrar a função "odbcConnectExcel2007"
File reading error
Error in inherits(channel, "RODBC") : objeto 'channel' não encontrado
GCDkit-> loadData("teste3.csv")
CSV file...loading...
...ok
teste3.csv
Error in kde[, f] : índice fora de limites
File reading error
Error in inherits(channel, "RODBC") : objeto 'channel' não encontrado
But as you suggested, copying with ctrl+c and pasting with ctrl+v was supposed to work? Because it is not. I'm getting error message of unexpected numeric constant for the numbers in the columns.
Is there a special command for "Paste Data from Clipboard" instead of ctrl+v?
Sorry, but I am new at not having the GUI.
Isabela
Dear Isabela,
Delete[1] You should be able to switch on the GUI on Mac by typing in the Console window:
menuet()
[2] Try to load the sample data file sazava.data
[3] No, Ctrl-V will not work, you have to invoke "Paste data from clipboard" from the menu
Please let me know, how you are getting on.
Vojtech
Hi Vojtech,
Delete[1] I was able o switch on the GUI with menuet(), but there is a message "Error in menuOff() : object 'schema.db' not found" once I do it . Options inside each tab of the GUI are not available to click. Only "Load data file" and "Paste data from clipboard". And there are error messages also when I try to use them. But...
[2]I successfully loaded the sazava.txt using the batch mode and then I managed to load my own data. I must have done something wrong in formatting tables the other day.
I will work with that! Thank you very much.
Hello Isabela,
Deleteyou may like to hear that the platform-independent GCDkit 6.0 has been just released. See http://blog.gcdkit.org/2019/08/gcdkit-60-has-been-unleashed-what-is-new.html?m=1
Ovulation Calendar is a easy and simple to use application to calculate the actual time of ovulation and determine the fertile period.
ReplyDeleteHi!
ReplyDeleteI am trying to install the 5.0 version on my win10 but when I load the package it always suggested that something is wrong with line 216 in"EarthChemFUN.r". I checked the file and find that there is a strange character under UTF-8 ("zzz=="something not correctly shown"", seems to be a \hat{A} but only shown under Windows-1252). I commented this line out and now it seems to work fine.
I am using the Chinese version of win10 with English language pack and this might cause the encoding problem. So I wonder if I need to install another language pack? Or commenting this line out would not affect the program?
Thanks!
Jiacheng Tian
Hello,
Deleteyou are not the first Chinese user reporting this problem with encoding in the EartChem module. But you can perhaps try us to test a remedy for us. Could you please replace the offending line of code in the file ErathChemFUN.r by:
if(.Platform$OS.type=="windows") zzz[zzz==intToUtf8(0x00c2)]<-NA
Please let me know whether it has worked. If so, we will add it to the next release.
Cheers,
Vojtech
Hi Vojtech,
DeleteAfter changing the code I could load gcdkit package and use it now. But as I don't use Earthchem module, I don't know whether it also works well after the change.
Thank you for your help!
Jiacheng Tian
Dear Jiacheng Tian,
DeleteI am pleased, thank you for letting me know.
Vojtech
how can I use Logarthmic axis please ?
ReplyDeleteSimply use the menu Plots|Binary plot, and when asked to specify which of the axes should be logarithmic, reply x, y, or xy. See also indication in the relevant dialogue box.
ReplyDeleteHello
ReplyDeleteI'd like to add a new normalization sceme to the spiderplots.
How can I do that using the 5.0 version?
It is the same like before, see
ReplyDeletehttp://blog.gcdkit.org/2013/06/how-to-introduce-your-own-normalization.html?m=1
Vojtech
Mr Janousek, thanks for replying.
DeleteI was having difficulties to find the spider.data file, but now I got it. Thank you again.
Hi every one. Im from Mexico City... In those days i have been triying to install the new version 5.0 on R 3.4.3. Im working whit ANACONDA R enviroment but i can't install. Some idea?...
ReplyDeleteThis comment has been removed by the author.
ReplyDeleteThis comment has been removed by the author.
ReplyDeleteHello!
ReplyDeleteIt was a good news for me that it now can be installed on Linux, because recently I moved from windows to Ubuntu and was missing GCDkit on it. So I followed the instruction and installed it. When I type library(GCDkit) in the R console, the GCDkit package loads, despite it can't read the config file: "Error reading the GCDkit config file!". However, the GUI does not load up. Reading the comments from others here, similar to Mac, I tried to load the GUI by typing menuet(), but there is no such function. I also tried to type menu(), but apparently the menu function needs an argument "choices", which I don't know what is.
Could you, please, point me in the right direction for loading the GUI on Ubuntu?
Thanks.
Best regards,
Jovid.
Dear Jovid,
DeleteI am sorry for your problems. As it has been mentioned in my post, the non-Windows GUI in GCDkit 5.0 is an undocumented feature meant for our testing only, and it turned out to have some serious problem.
But there is no need to despair, we are just finishing GCDkit 6.0, including some testing on Mac and Linux systems. It will contain tcl/tk GUI as one of the main attractions. It should be released before Goldschmidt in Barcelona (i.e. within a month), where I will have a presentation on platform-independent GCDkit.
Cheers,
Vojtech
Dear Vojtech,
DeleteThanks a lot for reply. I will be looking forward for the cross-platform GCDkit 6.0.
Best regards,
Jovid.
Dear Jovid and others,
Deletehere it is, platform-independent GCDkit has been released almost two weeks ago. See http://blog.gcdkit.org/2019/08/gcdkit-60-has-been-unleashed-what-is-new.html
Vojtech
Dear Vojtech,
ReplyDeleteI have been a GCDkit user for a long time. Over the past few months, I have noticed that the number of geotectonic models in the list has been decreased. I have tried to install a new version that is GCDkit 6.0 but I could not find the solution.
Looking forward to your reply, thanks.
Cheers
Dear Farhan,
ReplyDeletethank you for kind words. This sounds strange - in fact, the number of geotectonic plots increases all the time. Perhaps the problem is, that the list of available plots reflects the dataset loaded - if there are no data for the given diagram, it simply is omitted from the list.
Cheers,
Vojtech
hello
ReplyDeleteI drag the install file from GCd to R it shows file name desc error how can fix it.
ello
ReplyDeleteI drag the install file from GCd to R it shows file name desc error how can fix it.
Hello,
Deleteplease check first http://blog.gcdkit.org/2020/06/known-gcdkit-60-installation-problems.html
If the problems persist, please make a proper bug report with details of your OS, version of GCDkit, what have you done and what happened. I am not fortune teller ;-).
Best wishes,
Vojtech Janousek
If you require SEO services, Philadelphia seo is usually the best alternative. They will provide experienced services and a competent crew to accomplish all of their clients' assignments, and they will consistently deliver better outcomes and higher rankings.
ReplyDeleteI have GCDKit installed on two PC's, one with windows 8.1 and one with w10. Both worked till recently with the earthchem database interface. However, since some time now the R interface states that it couldn't find the EarthChemGUI when trying to launch it from the drop down menu. Any known issue or did I manage to crash two R + GCDkit installations?
ReplyDeleteHere is the detailed error print: Error in .EarthChemGUI() : could not find function ".EarthChemGUI"
They have changed the address of the EarthChem web service recently, try to install the newest version, just out.
ReplyDelete